In silico analysis
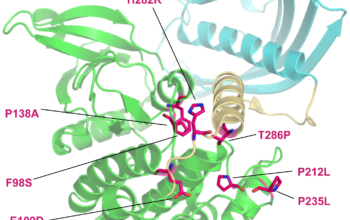
In Silico studies Mutation frequency and protein structure To assess the likelihood of pathogenicity, missense mutations are first screened against large databases such as the EXAC, SFARI and the ClinVar and LOVD database as well as in-house databases, To investigate how missense variants affect protein structure, we use prediction programs such as PolyPhen and SuSPect […]
Read moreIn vitro assays
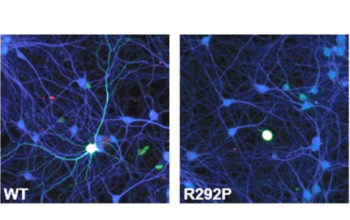
In vitro studies Protein stability and neuronal differentiation We use two in-vitro assays to assess the stability and pathogenicity of the mutation. The stability of the mutated protein is tested by expressing the protein in HEK cells. The pathogenicity of a mutation is tested by (over)expressing the protein in primary neuronal cultures. Common read-outs for […]
Read moreIn vivo assay
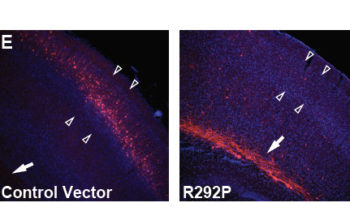
In vivo studies Neuronal migration For the primary in vivo assay of PRiSM, we have chosen to implement in utero electroporation (IUE), which allows us to quickly assess if a variant has a deleterious effect on in vivo neural progenitor cell (NPC) function, such that it interferes with neural migration to the cortical layers. […]
Read more